Research Summary
The Llobet-Navàs Laboratory conducts multidisciplinary research aimed at uncovering fundamental mechanisms driving gynecological cancers, with a particular emphasis on endometrial and breast tumors. Our projects involve close collaboration with computational biologists, clinical surgeons, pathologists, and medical oncologists at the Bellvitge University Hospital (HUB), which significantly enhances the translational potential of our findings. At the core of our work is a combination of molecular, biochemical, and cellular approaches, complemented by in vivo studies using genetically engineered mouse models and patient-derived xenografts.
We are currently pursuing several major research lines:
-
1. Anoikis Resistance in Gynecological Tumors
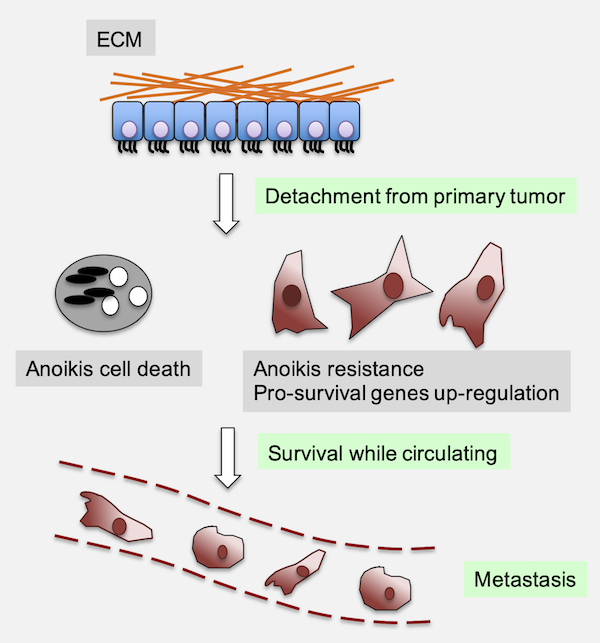
Fig.1 - Anoikis evasion contributes to the metastatic process. Anoikis is a specialized form of programmed cell death that occurs when epithelial cells lose their attachment to the extracellular matrix (ECM). Under physiological conditions, this mechanism serves as a critical safeguard against aberrant cell survival and ectopic growth, particularly in epithelial tissues. In cancer, however, the ability of tumor cells to resist anoikis is a key prerequisite for metastatic dissemination, allowing detached cells to survive in circulation, colonize distant tissues, and form secondary tumors.
Our laboratory has launched a dedicated project to systematically dissect the genetic and biochemical foundations of anoikis resistance in gynecological cancers. The primary goals of this initiative are to identify the key regulators and signaling networks that allow tumor cells to survive under anchorage-independent conditions and, ultimately, to exploit these vulnerabilities for therapeutic intervention and the prevention of metastasis.
To accomplish this, we are implementing a multi-tiered, systems-level strategy that integrates diverse experimental approaches. At the core of our discovery platform are genome-scale CRISPR interference (CRISPRi) loss-of-function screens conducted in three-dimensional (3D) suspension cultures, which enable us to pinpoint genes essential for cell survival in the absence of matrix attachment. These findings are complemented by transcriptomic profiling (RNA sequencing) and metabolomic analyses aimed at characterizing the global adaptations that support anoikis resistance, including the rewiring of metabolic circuits and activation of stress response pathways.
Following candidate gene identification, we perform functional validation using siRNA and shRNA knockdown strategies, as well as pharmacological inhibition, in both traditional monolayer cultures and physiologically relevant 3D models. In parallel, in vivo studies using orthotopic and metastatic mouse models allow us to evaluate the impact of candidate regulators on tumor dissemination, colonization, and response to therapy. Finally, to ensure clinical relevance, we incorporate patient-derived tumor samples and associated clinical metadata to assess the prognostic and predictive value of anoikis-related gene signatures, with particular attention to their role in therapeutic resistance and disease recurrence.
-
2. Autophagy in Endometrial Cancer Progression
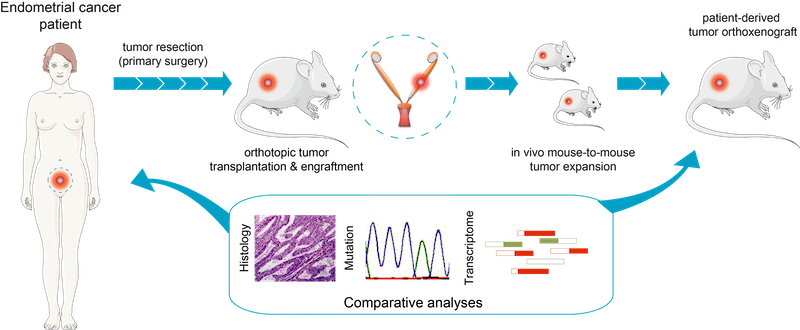
Fig.2 - Generation of EC orthoxenografts. Schematic representation of patient-derived orthoxenotransplant implants procedure and treatment used in Eritja et al. Autophagy 2017. EEC tumors were surgically removed and small pieces were implanted in the uterus of recipient female mice. Once engrafted, tumors were propagated to a cohort of 20–45 mice, randomized and treated accordingly. Endometrial cancer (EC) is the most common gynecological malignancy, with a subset of patients (~15–20%) experiencing recurrence or metastasis despite standard treatment. Unfortunately, systemic therapies offer limited benefit, highlighting the urgent need for novel targeted interventions.
Our laboratory has identified autophagy as a key process contributing to therapy resistance in EC (Eritja et al., Autophagy 2017; Devis-Jauregui et al., Autophagy 2021). We are now investigating the molecular drivers and regulatory networks that govern autophagy in endometrial tumor cells. By combining high-content fluorescence microscopy with autophagic flux assays in 3D EC cell cultures and patient-derived organoids (PDOs), we aim to identify druggable components of the autophagy machinery. These findings will be tested in vivo using our unique and continuously growing collection of orthotopic EC xenograft models (n > 150), in collaboration with clinical partners.
-
3. Targeting novel vulnerabilities in endometrial cancer through autophagy-related drug discovery
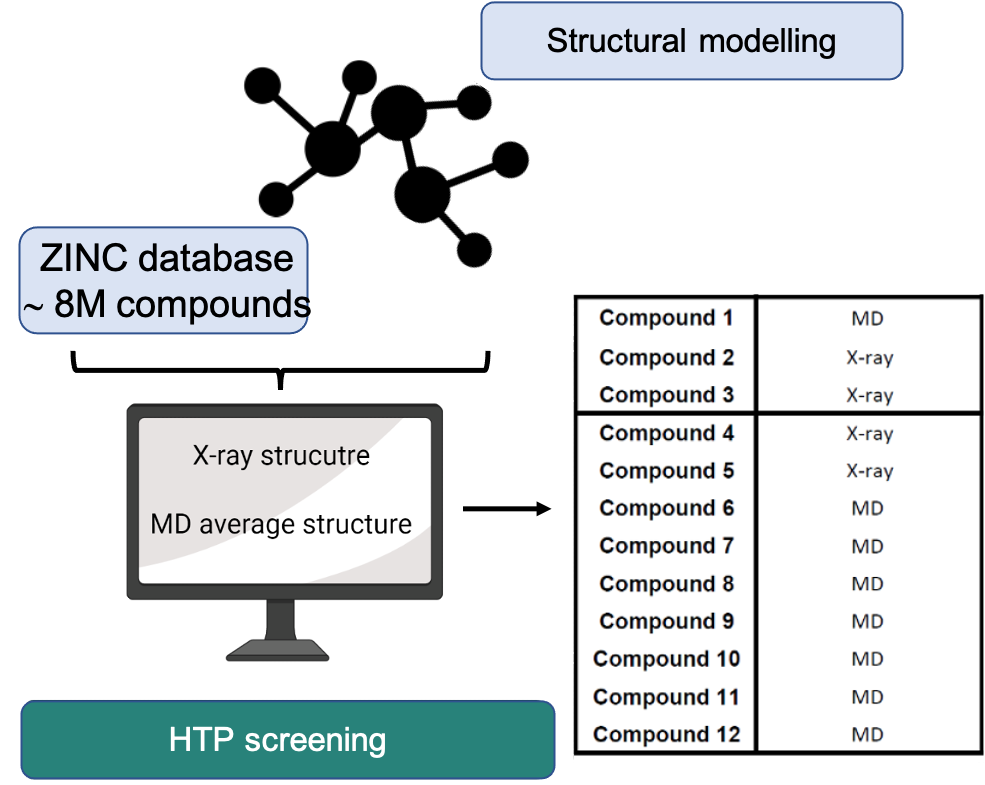
Fig.3 - High-throughput virtual screening and structural modeling pipeline for autophagy-targeted drug discovery in endometrial cancer. Over the past several years, our laboratory has concentrated on understanding how cellular self-digestion mechanisms, namely autophagy, contribute to cancer cell survival and therapy resistance. Building on this foundation, we recently developed and implemented a gene discovery platform focused on identifying druggable autophagy regulators with potential roles in endometrial cancer (EC) pathogenesis. Through this approach, we uncovered a previously unexplored molecular vulnerability that can be pharmacologically targeted. Using a clinically approved drug as a structural blueprint, we launched a large-scale in silico screening campaign involving over 8 million compounds. This strategy has yielded a number of promising small molecule candidates with the potential to interfere with this newly identified target. Our current efforts focus on the chemical optimization of these candidates, guided by advanced structural modeling and high-precision protein-drug interaction analyses. These include assessments of binding kinetics, affinity, and specificity. Concurrently, we are conducting comprehensive in vitro and in vivo evaluations to determine their therapeutic efficacy against EC. The ultimate goal of this project is to develop a novel, potent, and safe targeted therapy for endometrial cancer, one that could offer a much-needed precision medicine option for patients who currently have limited alternatives.
-
4. Investigating microRNA-mediated control of oncogenic signaling in endometrial cancer
Our laboratory is committed to understanding how microRNAs (miRNAs) contribute to the development and progression of endometrial cancer (EC), particularly in tumors characterized by dysregulated PI3K/AKT and TGFβ signaling, two pathways frequently altered in this disease. Despite their known roles in promoting proliferation and suppressing apoptosis, the regulatory elements that modulate their output in the endometrial epithelium remain incompletely defined. In this project, we aim to investigate how specific miRNA programs act as integrators of oncogenic and tumor-suppressive signaling in EC. We hypothesize that certain miRNA clusters, regulated by upstream signaling dynamics, may control key transcriptional networks involved in cellular proliferation, survival, and apoptotic resistance. To address this, we will employ a combination of in vitro and in vivo approaches, including genetically engineered mouse models, 3D endometrial organoid cultures, transcriptomic profiling, and targeted functional studies. These complementary strategies will allow us to dissect the mechanistic role of miRNAs in fine-tuning the cellular response to oncogenic stress and in shaping the trajectory of tumor progression.
-
5. miRNAs in Breast Tumorigenesis and Stemness
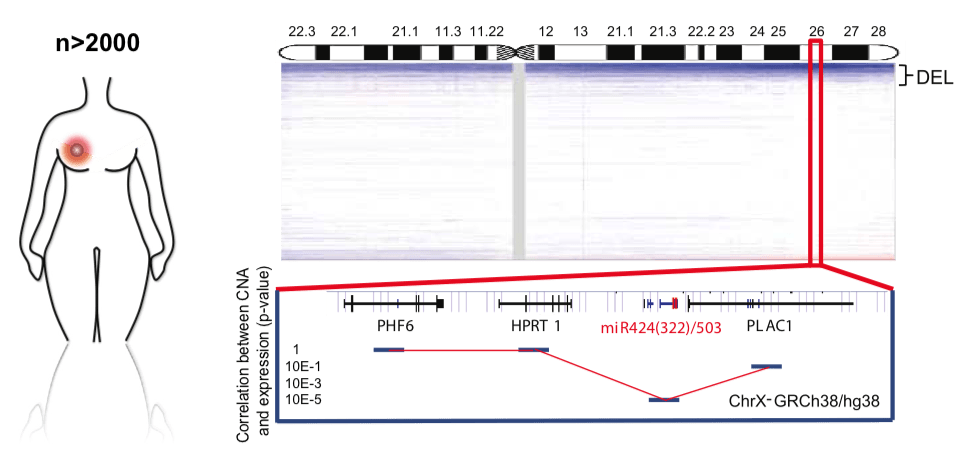
Fig.5 - miR-424/503 is lost in breast cancer and is associated with poor prognosis and aggressive subtypes. Copy number profile of the X chromosome in human breast cancers (METABRIC plus TCGA data sets), highlighting the chromosomal region containing the miR-424/503 locus. The correlation between copy number and expression for surrounding genes is included at the bottom. We are investigating how the loss of specific microRNA clusters influences stemness and tumor development in the mammary gland. Our earlier work identified the miR-424/503 cluster as a key regulator of mammary gland involution (Llobet-Navas et al., Genes & Development 2014; Molecular and Cellular Biology 2014), under transcriptional control of the TGFβ pathway. Mechanistically, this cluster modulates apoptosis and cell cycle regulation via CDC25A, BCL2, and IGF1R.
In collaboration with clinical and genomic researchers, we found that approximately 15% of breast tumors harbor deletions in this miRNA cluster (Rodriguez-Barrueco et al., Genes & Development 2017) and present increased Wnt/B-catenin activity (Nekritz et al., EMBO Reports 2021). Genetically engineered miR-424/503-deficient mice spontaneously develop mammary tumors resistant to standard chemotherapies; however, these tumors show sensitivity to IGF1R and BCL2 inhibitors currently in clinical trials. Our ongoing efforts include characterizing the genetic and transcriptomic profiles of these tumors and identifying their cell-of-origin.
-
6. miRNAs in Adipose Tissue Biology
Fig.6 - Representative illustration of the project. Building on our in vivo data (Rodriguez-Barrueco et al., Adv Sci 2022), we are investigating the role of the miR-424/503 cluster in adipocyte differentiation and adipose tissue (AT) homeostasis. This line of research focuses on dissecting the molecular and metabolic consequences of miR-424/503 loss through the use of knockout mouse models. Specifically, we aim to characterize the obese phenotype observed in miR-424/503-deficient mice and to define the miRNA regulon that governs adipocyte differentiation. In parallel, we are correlating miRNA expression levels in human adipose tissue samples with clinical outcomes, and examining genetic polymorphisms that may influence the expression of this miRNA cluster. Collectively, these studies are designed to provide new insights into the regulation of adipose tissue expansion and its broader implications for metabolic disease and cancer.
Our research is or has been funded by






